Contact Us
For additional information about the Computational Lung Physiology Laboratory, Contact the CLPL.
Report a Problem
To report a problem with this website, contact BME Communications or report an accessibility issue.
Lung diseases and injuries are frequently initiated by or are a consequence of changes in lung tissue bioenergetics, the biochemistry of how cells utilize and transform energy. A wealth of information exists regarding the bioenergetic processes in mitochondria obtained from lungs and cultured cells under physiological and pathophysiological conditions; however, the interdependence of those processes makes it difficult to quantify the impact of a change in a single or multiple process(es) on overall lung tissue bioenergetics. Integrated computational modeling provides a mechanistic and quantitative framework for the bioenergetic data at different levels of biological organization.
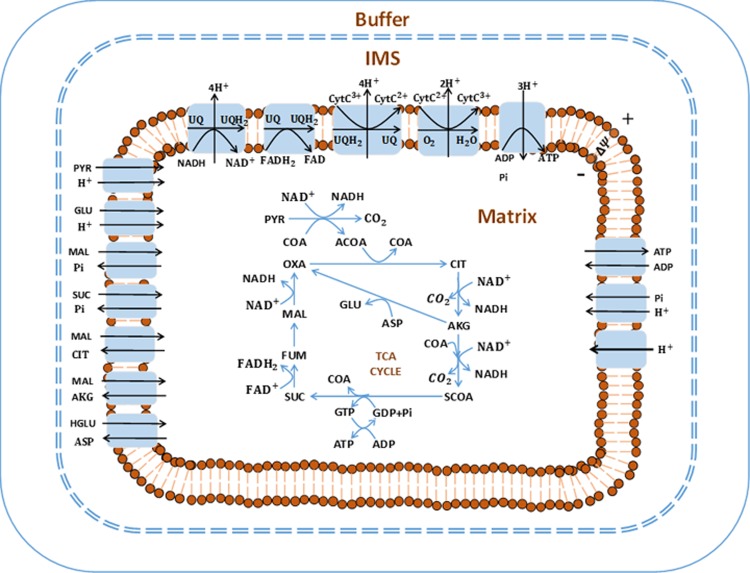 We have developed a model of the bioenergetics—including major biochemical reactions and transport processes—of lung mitochondria. Intrinsic model parameters, such as binding constants, are estimated using enzyme and transporter kinetic data. Extrinsic model parameters, such as maximal reaction and transport velocities, are estimated by fitting the model to tricarboxylic acid cycle and respirometry data measured in rat lung mitochondria. The model is validated by assessing its ability to predict experimental data not used in the estimation process. This work has been extended to a complex model of whole-lung bioenergetics. This model includes both mitochondrial and cytosolic regions and accounts for glucose uptake and phosphorylation, glycolysis, and the pentose phosphate pathway. This computational modeling provides a mechanistic and quantitative framework for integrating lung tissue bioenergetics data from various sources and for testing novel hypotheses regarding the role of different cytosolic and mitochondrial processes in lung tissue bioenergetics. For example, we have used the model to gain novel insights on how lung tissue glycolytic rate is regulated by exogenous substrates such as glucose and lactate, which might be affected with lung injury or disease.
We have developed a model of the bioenergetics—including major biochemical reactions and transport processes—of lung mitochondria. Intrinsic model parameters, such as binding constants, are estimated using enzyme and transporter kinetic data. Extrinsic model parameters, such as maximal reaction and transport velocities, are estimated by fitting the model to tricarboxylic acid cycle and respirometry data measured in rat lung mitochondria. The model is validated by assessing its ability to predict experimental data not used in the estimation process. This work has been extended to a complex model of whole-lung bioenergetics. This model includes both mitochondrial and cytosolic regions and accounts for glucose uptake and phosphorylation, glycolysis, and the pentose phosphate pathway. This computational modeling provides a mechanistic and quantitative framework for integrating lung tissue bioenergetics data from various sources and for testing novel hypotheses regarding the role of different cytosolic and mitochondrial processes in lung tissue bioenergetics. For example, we have used the model to gain novel insights on how lung tissue glycolytic rate is regulated by exogenous substrates such as glucose and lactate, which might be affected with lung injury or disease.
Integrated Computational Model of Lung Tissue Bioenergetics. (Zhang X, Dash RK, Clough AV, Xie D, Jacobs ER, Audi SH) Front Physiol 2019;10:191 PMID: 30906264
Integrated Computational Model of the Bioenergetics of Isolated Lung Mitochondria. (Zhang X, Dash RK, Jacobs ER, Camara AKS, Clough AV, Audi SH) PLoS One 2018;13(6):e0197921 PMID: 29889855
The Computational Lung Physiology Laboratory has a strong commitment to training the next generation of scientists interested in using state-of-the-art computational techniques to improve human health. Research opportunities are available for undergraduate and graduate (MS and PhD) students alike. For more information on joining our team, Contact the CLPL !