Contact Us
For additional information about the Computational Lung Physiology Laboratory, Contact the CLPL.
Report a Problem
To report a problem with this website, contact BME Communications or report an accessibility issue.
The Computational Lung Physiology Laboratory (CLPL) has facilities located at both the Marquette University and Zablocki VA Medical Center campuses. These laboratories are equipped to facilitate a wide range of investigative and educational activities, with special emphasis placed on molecular imaging and computational modeling. In addition, the facilities feature optical imaging modalities, various ventilation-perfusion systems, animal models of clinical lung disease and other amenities afforded by collaborations with various laboratories of both Marquette University and the Medical College of Wisconsin.
 The CLPL is currently using different biomarkers with SPECT/CT imaging to measure regional blood flow, cell death, mitochondrial function, and tissue redox status. The overall approach is to administer a tagged biomarker that accumulates in the tissue or organ of interest and to noninvasively detect that biomarker with appropriate imaging cameras. The imaging data is then analyzed using pharmacokinetic modeling to identify and quantify a targeted process.
The CLPL is currently using different biomarkers with SPECT/CT imaging to measure regional blood flow, cell death, mitochondrial function, and tissue redox status. The overall approach is to administer a tagged biomarker that accumulates in the tissue or organ of interest and to noninvasively detect that biomarker with appropriate imaging cameras. The imaging data is then analyzed using pharmacokinetic modeling to identify and quantify a targeted process.
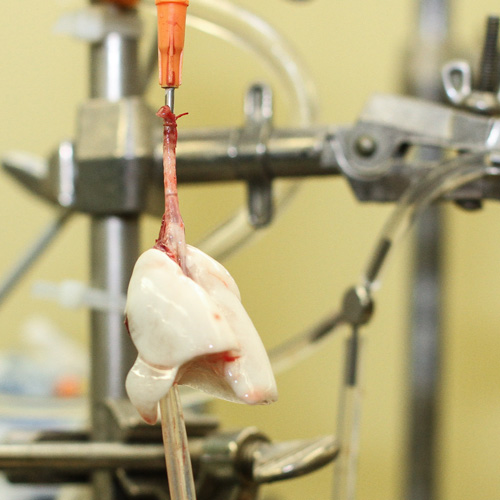 Investigators at the CLPL use isolated perfused lung (IPL) model systems for rodents. In this process, the lung is connected to a mechanical ventilator and pump perfusion system that allows for external regulation or respiration and blood flow. Lung hemodynamics and cellular function can be manipulated by changing respiration, pump flow, vascular pressures, and/or introducing compounds into the inflow to the lung. IPL systems with accompanying equipment are currently being used to perform molecular and optical imaging, to obtain global measurements of cellular functions (including oxidative stress and mitochondrial function), and to measure vascular permeability under normal and pathophysiologic conditions.
Investigators at the CLPL use isolated perfused lung (IPL) model systems for rodents. In this process, the lung is connected to a mechanical ventilator and pump perfusion system that allows for external regulation or respiration and blood flow. Lung hemodynamics and cellular function can be manipulated by changing respiration, pump flow, vascular pressures, and/or introducing compounds into the inflow to the lung. IPL systems with accompanying equipment are currently being used to perform molecular and optical imaging, to obtain global measurements of cellular functions (including oxidative stress and mitochondrial function), and to measure vascular permeability under normal and pathophysiologic conditions.
 The CLPL has two fluorescence imaging systems (Photon Technology International, HORIBA Scientific) and access to additional systems for optical imaging of isolated perfused lungs, tissue samples, cultured cells, or isolated organelles including mitochondria. These systems can be used to measure oxygen consumption, respiratory control index (mitochondrial membrane potential), and H2O2 formation as an index of oxidative stress, for example.
The CLPL has two fluorescence imaging systems (Photon Technology International, HORIBA Scientific) and access to additional systems for optical imaging of isolated perfused lungs, tissue samples, cultured cells, or isolated organelles including mitochondria. These systems can be used to measure oxygen consumption, respiratory control index (mitochondrial membrane potential), and H2O2 formation as an index of oxidative stress, for example.
 The CLPL has state-of-the-art equipment for measuring breathing rate and dynamic lung compliance via plethysmography and blood oxygenation with pulse oximetry. Standard monitoring of heart rate, blood pressure, etc., can also be performed in vivo. Moreover, the lab performs an extensive list of tissue assays, including histological and biomolecular analyses for oxidative stress, inflammation, cell death, etc.
The CLPL has state-of-the-art equipment for measuring breathing rate and dynamic lung compliance via plethysmography and blood oxygenation with pulse oximetry. Standard monitoring of heart rate, blood pressure, etc., can also be performed in vivo. Moreover, the lab performs an extensive list of tissue assays, including histological and biomolecular analyses for oxidative stress, inflammation, cell death, etc.
 Investigators at the CLPL have extensive experience with lung-injury models designed to replicate key features of sepsis, ARDS, and other pulmonary vascular disease injuries. These include chronic models involving hypoxic and hyperoxic exposure chambers, as well as tracheal and IP administration of endotoxins. Investigators also utilize techniques designed to induce acute injury by modulating vasoreactivity, mitochondrial function, and cellular metabolism. Moreover, the team has access to a wide array of genetically modified rodents through the Medical College of Wisconsin.
Investigators at the CLPL have extensive experience with lung-injury models designed to replicate key features of sepsis, ARDS, and other pulmonary vascular disease injuries. These include chronic models involving hypoxic and hyperoxic exposure chambers, as well as tracheal and IP administration of endotoxins. Investigators also utilize techniques designed to induce acute injury by modulating vasoreactivity, mitochondrial function, and cellular metabolism. Moreover, the team has access to a wide array of genetically modified rodents through the Medical College of Wisconsin.
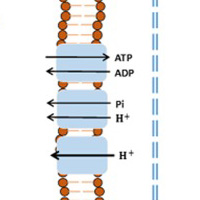 Physiologically-based pharmacokinetic modeling of experimental data is used extensively to identify and quantify changes in key cellular processes hypothesized to be involved in lung injury. The models account for lung tissue vascular permeability, capillary perfusion heterogeneity, cellular reactions, blood, and the systemic circulation on the lung uptake of the relevant probes or biomarkers. These models provide an integrative mechanistic approach for proper interpretation of in vivo and isolated perfused lung data.
Physiologically-based pharmacokinetic modeling of experimental data is used extensively to identify and quantify changes in key cellular processes hypothesized to be involved in lung injury. The models account for lung tissue vascular permeability, capillary perfusion heterogeneity, cellular reactions, blood, and the systemic circulation on the lung uptake of the relevant probes or biomarkers. These models provide an integrative mechanistic approach for proper interpretation of in vivo and isolated perfused lung data.
The Computational Lung Physiology Laboratory has a strong commitment to training the next generation of scientists interested in using state-of-the-art computational techniques to improve human health. Research opportunities are available for undergraduate and graduate (MS and PhD) students alike. For more information on joining our team, Contact the CLPL !