Graduate Student Research
Biomedical Engineering Posters | Awards
Graduate students of the Marquette University and Medical College of Wisconsin Joint Department of Biomedical Engineering contribute to ongoing research projects in an array of disciplines. Working hand-in-hand with our world-class faculty, students engage in cutting-edge research in their work to advance the knowledge base and translational investigations at the heart of scientific advancement.
Biomedical Engineering Posters
 Blood Outgrowth Endothelial Cell Response to Culture on Novel Ferromagnetic Flow Diverting Stent
Blood Outgrowth Endothelial Cell Response to Culture on Novel Ferromagnetic Flow Diverting Stent
Presented by Aleksandra Zielonka under the direction of Dr. Brandon J. Tefft
-
Poster (Click to open as PDF)
-
Abstract
-
Authors
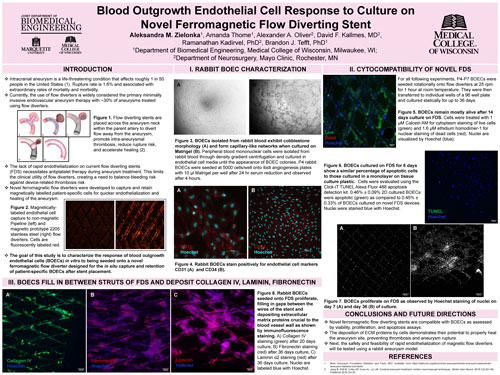
When flow diverters are introduced to the site of an aneurysm, endothelial cells must adhere and cover the aneurysm neck for successful long term aneurysm occlusion. The current lack of rapid endothelialization on flow diverting stents necessitates antiplatelet therapy during aneurysm treatment with flow diverting stents. This limits the clinical utility of flow diverters, creating a need to balance bleeding risk against device-related thrombosis risk. This study characterized the response of blood outgrowth endothelial cells (BOECs) in vitro to being seeded onto a novel ferromagnetic flow diverter designed for the in situ capture and retention of patient-specific BOECs after stent placement. Peripheral blood mononuclear cells were isolated from rabbit blood and cultured in endothelial cell media until the appearance of BOEC colonies. BOEC phenotype was confirmed using tube formation assay and immunofluorescence with CD31 and CD34. BOECs were rotationally seeded onto devices and cultured statically inside wells of a 96 well plate. No notable difference in cell proliferation, apoptosis, and viability was observed compared to controls. Immunofluorescence using antibodies against collagen IV, laminin, and fibronectin revealed extracellular matrix (ECM) deposition by BOECs seeded onto the flow diverting stent. Importantly, cells and ECM were found both on and between the wires of the device. The deposition of ECM proteins by cells may demonstrate their potential to properly heal the aneurysm site, preventing thrombosis and aneurysm rupture.
Aleksandra M. Zielonka, Amanda Thome, Alexander A. Oliver*, Ramanathan Kadirvel*, David F. Kallmes*, Brandon J. Tefft
 Towards Efficient Blood Outgrowth Endothelial Cell Seeding of small diameter Gore-Tex Vascular Graft
Towards Efficient Blood Outgrowth Endothelial Cell Seeding of small diameter Gore-Tex Vascular Graft
Presented by Aysan Hedayatnazari under the direction of Dr. Brandon J. Tefft
-
Poster (Click to open as PDF)
-
Abstract
-
Authors
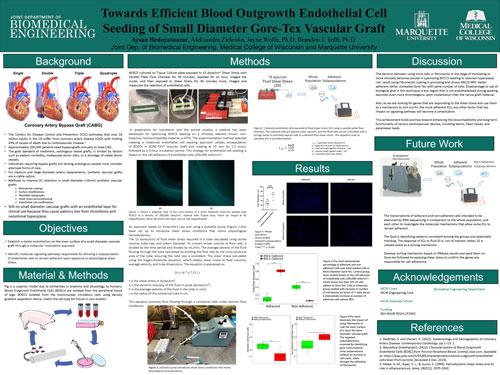
Cardiovascular disease, a leading cause of morbidity and mortality in developed nations, necessitates innovative solutions for hemocompatibility in small-diameter vascular grafts.
The need for progress is emphasized by the lack of a synthetic biomaterial considered safe for vessels smaller than 6mm. This project aims to establish a stable endothelium on implantable cardiovascular devices through a molecular modulation approach. Overcoming barriers, the research investigates a subpopulation of endothelial cells resistant to detachment, uncovering crucial molecular signaling pathways. Optimizing cell seeding on a known non-bioactive material and introducing shear stress conditions mimicking physiological hemodynamics proved to be essential. Achieving a seeding density of 200,000 endothelial cells on Gore-Tex marked a significant success, laying the foundation for investigating adherent cell sorting and signaling pathways under shear stress. This achievement holds promise for enhancing the biocompatibility and long-term functionality of various cardiovascular devices, including stents, heart valves, and pacemaker leads. The decision between using more cells or fibronectin becomes pivotal in optimizing these devices for reduced hyperplasia risk.
Aysan Hedayatnazari, Aleksandra Zielonka, Jayne Wolfe, Brandon J. Tefft
 Estimating the dose of dry powder inhalers delivered to the larynx using computational fluid dynamics
Estimating the dose of dry powder inhalers delivered to the larynx using computational fluid dynamics
Presented by Shamudra Dey under the direction of Dr. Guilherme Garcia
-
Poster (Click to open as PDF)
-
Abstract
-
Authors
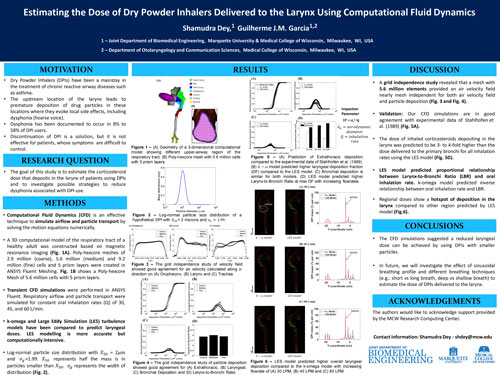
Inhaled corticosteroids are a mainstay in the treatment of asthma. However, corticosteroid deposition in the larynx elicits local side effects, including dysphonia (hoarse voice). The goal of this study was to estimate the corticosteroid dose that deposits in the larynx in patients using dry powder inhalers (DPIs) and to investigate possible strategies to reduce dysphonia associated with DPI use. A 3-dimensional computational model of the respiratory tract of a healthy adult was constructed based on magnetic resonance imaging. Respiratory airflow and particle transport were simulated using computational fluid dynamics (CFD) for oral inhalation rates of 30, 45, and 60 L/min. Log-normal particle size distributions with mass median aerodynamic diameters in the 1-50 microns range were investigated. The dose of inhaled corticosteroids depositing in the larynx was predicted to be 3- to 4-fold higher than the dose delivered to the primary bronchi for all inhalation rates. The CFD simulations suggested a reduced laryngeal dose can be achieved by inhaling the drug at a lower flow rate or using DPIs with smaller particles.
Shamudra Dey and Guilherme J.M. Garcia
 Limitations of intra-hemispheric exchange of attention information in PPC during shifts of object-based attention
Limitations of intra-hemispheric exchange of attention information in PPC during shifts of object-based attention
Presented by David Hughes under the direction of Dr. Adam Greenberg
-
Poster (Click to open as PDF)
-
Abstract
-
Authors
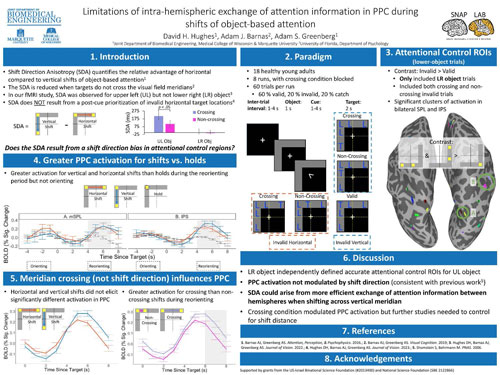
We have previously shown (Barnas & Greenberg, 2016, 2019), behaviorally, that shifts of object-based attention are asymmetric: there’s a horizontal (vs. vertical) shift advantage (the Shift Direction Anisotropy; SDA) which is larger when shifting attention across the visual field meridians. Here, we used fMRI in 19 healthy volunteers to investigate whether meridian crossing and shift direction modulate attention reorienting signals in posterior parietal cortex (PPC), a critical node controlling attention shifts. We hypothesized that, when subjects shift across the meridians, we would observe differences between horizontal and vertical reorienting signals indicating more efficient reallocation of attention during horizontal shifts (Hughes et al., VSS2022). Subjects were endogenously cued to attend the vertex of an L-shaped object (at one of 2 screen locations) and performed a target detection task with 5 possible target locations: (1) at the object vertex (valid); on the horizontal object arm (invalid-horizontal) at either a (2) near, non-crossing or (3) far, meridian crossing location; on the vertical object arm (invalid-vertical) at either a (4) near, non-crossing or (5) far, meridian crossing location. We used trials from one object location to identify regions of interest in PPC and extracted event-related averages time-locked to target onset for the other object. Results yielded greater activation on invalid (vs valid) trials (4 to 7 s; ts > 2.9, ps < 0.005) but not during the initial (cue-related) orienting (-4 to -1 s; ts < 1.4, ps > 0.15), confirming an expected validity effect. A repeated measures ANOVA with crossing condition (crossing, non-crossing), shift direction (horizontal, vertical), and time (4, 5, 6, 7 s) as within-subjects factors revealed all significant main effects (crossing condition: F(1, 18) = 30.0, p < 0.001; shift direction: F(1, 18) = 8.1, p < 0.005; time: F(3, 18) = 32.9, p < 0.001). The crossing X shift direction interaction was also significant, F(1, 18) = 4.9, p = 0.028. Post-hoc comparisons revealed greater activation during all crossing trials (vs. non-crossing), t(18) = 2.5, p = 0.023, d = 0.67. However, no crossing trial timepoints showed a significant difference between horizontal and vertical shifts (all ps > 0.60). Thus, while meridian crossing condition influenced PPC, there was no difference between horizontal and vertical reorienting across the meridians. We, therefore, conclude that the SDA arises not from a shift direction bias in PPC but, instead, via a more efficient exchange of information between (vs. within) hemispheres during horizontal (vs vertical) shifts, reflecting attentional resource limitations.
David Hughes, Adam J. Barnas*, Adam S. Greenberg
Awards
Every year, MCW's School of Graduate Studies and the Office of Postdoctoral Education's annual Research Poster Session evaluates graduate students and postdoctoral researchers on the quality of their research, poster design presentation. Winners are selected from both graduate student and postdoctoral fellow entrants.
The winners of this year’s graduate school poster session included Joint Department student Shamudra Dey for "Estimating the dose of dry powder inhalers delivered to the larynx using computational fluid dynamics."
Congratulations Shamudra!
Learn more about BME Research

 Blood Outgrowth Endothelial Cell Response to Culture on Novel Ferromagnetic Flow Diverting Stent
Blood Outgrowth Endothelial Cell Response to Culture on Novel Ferromagnetic Flow Diverting Stent
 Towards Efficient Blood Outgrowth Endothelial Cell Seeding of small diameter Gore-Tex Vascular Graft
Towards Efficient Blood Outgrowth Endothelial Cell Seeding of small diameter Gore-Tex Vascular Graft
 Estimating the dose of dry powder inhalers delivered to the larynx using computational fluid dynamics
Estimating the dose of dry powder inhalers delivered to the larynx using computational fluid dynamics
 Limitations of intra-hemispheric exchange of attention information in PPC during shifts of object-based attention
Limitations of intra-hemispheric exchange of attention information in PPC during shifts of object-based attention
